Pseudomonas is a gram-negative bacteria commonly found in soil, sediments, and freshwater. This bacteria was previously found in sub-alpine Himalayan forests and temperate Himalaya by Teramoto and colleagues (2007).
References
- Teramoto K, Sato H, Sun L, Torimura M, Tao H, Yoshikawa H, Hotta Y, Hosoda A, Tamura H. 2007. Phylogenetic classification of pseudomonas putida strains by maldi-ms using ribosomal subunit proteins as biomarkers. Anal Chem [Internet]. [cited 6 April 2017];79(22):8712-8719. Available from: http://pubs.acs.org/doi/pdf/10.1021/ac701905r
Date Collected: February 9, 2017
Methods of Isolation and Identification:
- Fifty milliliters of surface water was collected from Buffalo Creek (Figure 1). A serial dilution technique was used to create 3 nutrient agar plates. A sterile hockey stick was used to spread the water samples around the agar plates. These plates were then incubated at 25 degrees Celsius over an entire week (Thursday-Thursday).
- A light yellow colony (Figure 2) was selected for 16s rRNA gene sequencing by PCR amplification.
- The PCR product was digested with MspI and sequenced to identify the genus and species of the bacteria.


Figure 1. Site of collection – Buffalo Creek
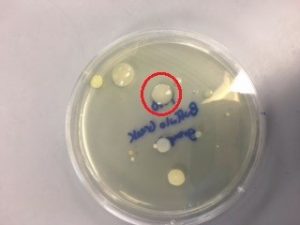
Figure 2. Colony selected for identification – from 1:10 dilution
Results:
- MspI Digestion (Figure 3): A 1,500 bp product was amplified using PCR. After digestion with MspI, two bands located at approximately 500 bp and 100 bp were identified.
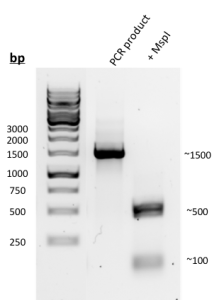
Figure 3. Results of PCR amplification and MspI digestion
- Sequence analysis (Figure 4 and 5): The sequences PCR product generated 638 bases of high-quality reads that were used to identify the genus of the colony. The chromatogram of the sequence is available as a pdf (8B_PREMIX_JF7514_10.pdf). NCBI BLAST analysis revealed a 0.63 difference between the identities of the 5 different Pseudomonas species (Figure 4). As an example, the top hit (99.53% – mismatched with only 3 bases) identified with bases 90-727 of the 16s rRNA gene of Pseudomonas putida. (Figure 5).
| Genus | Species | Strain | Identity | Accession # |
| Pseudomonas | putida | NBRC 14164 | (635/638) 99.53% | NR_113651.1 |
| Pseudomonas | corrugate | N/A | (632/638) 99.06% | NR_117826.1 |
| Pseudomonas | baetica | a390 | (631/638) 98.90% | NR_116899.1 |
| Pseudomonas | fuscovaginae | ICMP 5940 | (631/638) 98.90% | NR_116700.1 |
| Pseudomonas | kilonensis | 520-20 | (631/638) 98.90% | NR_028929.1 |
Figure 4. NCBI BLAST – top 5 results
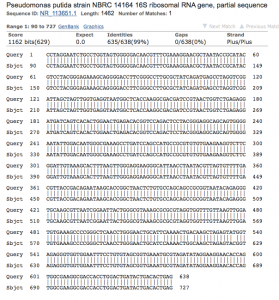
Figure 5. NCBI BLAST analysis of colony 8B
Contributed by: Caitlin Harris, Jessica Mick, and Kayla Cooksey, BIOL 250 Spring 2017, Group 8