Pseudomonas lurida strain P513/18 16s ribosomal
- Pseudomonas lurida is a microbial community that is a non-spore-forming rods that occurs as single cell and is gram negative (2007). It is known for its ability to produce a pigment showing a light yellow–green fluorescence by irradiation with UV-light at 350 nm (2007). This was found by a study. The type strain, P 513/18 was isolated from the phyllosphere of grasses in Paulinenaue, Germany (2007).
References: Behrendt, U., Ulrich, A., Schumann, P., Meyer, J. M., & Spröer, C. (2007). Pseudomonas lurida sp. nov., a fluorescent species associated with the phyllosphere of grasses. International journal of systematic and evolutionary microbiology, 57(5), 979-985.
Data Collected: March 31, 2017
Methods for isolation and identification:
- A stagnant surface water sample was taken from Buffalo Creek with a fifty millimeter conical tube (Figure 1). The direct count, 1:10, and 1:100 were created and placed into separate labeled agar plates with nutrient broth . They were then incubated for four days at 25 degrees Celsius where their colonies were established and grew.
- A white-yellowish, smooth, and round colony with regular margins was selected for 16S ribosomal RNA gene sequencing by polymerase chain reaction amplification (Figure 2).
- The polymerase chain reaction product was digested with the use of MspI and partially sequenced to identify the genus and then species of the bacteria
 Figure 1: Cite of Collection
Figure 1: Cite of Collection

Figure 2: Colony selected of Identification
Results:
MspI digestion (Figure 3): A 1,006 bp product was amplified by polymerase chain reaction where partial gene sequence was used on 800 bp. When digestion was done with MspI, two bands at approximately 250 bp and two bands around 500 bp were identified.
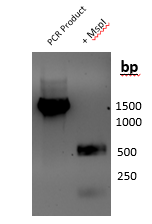
Figure 3: Results of PCR amplification and MspI digestion
Sequence analysis (Figure 4): When the polymerase chain reaction product was sequenced it produced 1,006 bases of high-quality reads that were compared. The chromatogram of the sequence is available as a pdf (
 Loading...
Loading...
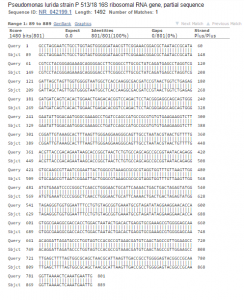 Figure 4: NCBI BLAST analysis of colony 16s
Figure 4: NCBI BLAST analysis of colony 16s
Contributed by: Victoria A. Acosta and Kaitlyn Marston, BIOL 250 Spring 2017, Group 3