Janthinobacterium lividum is a rod-shaped, Gram-negative, motile, aerobic bacterium, commonly isolated from of soil and water of cold regions. Janthinobacterium lividum colonizes the skin of some amphibians and confers protection against fungal pathogens.
Citation:
Valdes, N., Soto, P., Cottet, L., Alarcon, P., Gonzalez, A., Castillo, A., … Tello, M. (2015). Draft genome sequence of Janthinobacterium lividum strain MTR reveals its mechanism of capnophilic behavior. Standards in Genomic Sciences, 10, 110. doi: 10.1186/s40793-015-0104-z
Date Collected: February 8, 2017
Methods for Isolation & Identification:
- Collection and Sampling of Environmental Samples at Environmental Education Center
- For each collection site, we labeled 3 nutrient agar plates as follows: “direct count,” “1:10,” & “1:100“
- To plate these samples, we used the serial dilution technique
- Analyzing Data
- After 7 days, we collected the following data for each plate: number of colonies, color, size, shape, texture, form, elevation, and margin.
- Genomic DNA Isolation/Extraction
- Genomic DNA was extracted from two colonies to identify the bacterial species.
- Polymerase Chain Reaction (PCR)
- The 16s rDNA sequence of our own two unknown bacteria was amplified.
- Restriction Enzyme Digestion & Gel Electrophoresis
- The purpose of this step was to cleave/ligate a functional piece of DNA predictably and precisely (Restriction Enzyme Digestion).
- After cleavage, DNA fragments was separated using agarose gel electrophoresis.
- These steps will result in the DNA sequencing step.
- DNA Sequencing Analysis/Identification
- The purpose of this step was to identify our prokaryotes based on the sequence of the 16s rDNA. Also, analyzing our DNA sequences and identifying what prokaryote we likely identified.
- SnapGene viewer to analyze our sequence, used a BLAST to identify the prokaryote, and checked the MspI digestion sites for our sequence using NEB cutter
Results:
The sequenced PCR product for the 16s rRNA gene generated 857 bases of high-quality reads that were used to identify the genus and species of the bacteria colonies. This allowed us to separate and analyze the DNA and their fragments, based on their size and charge. The Gel Electrophoresis image below shows the nucleic acid molecules separated by bands. The shorter molecules moved faster and traveled farther than the longer ones because the shorter molecules traveled through the pores of the gel. As you can clearly see in the image below bands lining up, which represents a protein that compromises that band is highly abundant.
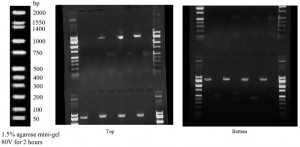
The digested gels are 3, 5, 7, and 9 with the top being Buffalo Creek tree at 5m, tree at 10m, and the two water samples.
Sequence Analysis:
The sequenced PCR product generated 865 bases of high-quality reads that were used to identify the genus and species of the colony. The chromatogram of the sequence is available as a pdf (BCW1) . NCBI BLAST analysis revealed 99% identity with bases 28-892 of the 16s rRNA gene of Janthinobacterium lividium.
Janthinobacterium lividum strain DSM 1522 16S ribosomal RNA gene, partial sequence
| Score | Expect | Identities | Gaps | Strand |
|---|---|---|---|---|
| 1585 bits(858) | 0.0 | 863/865(99%) | 1/865(0%) | Plus/Plus |
Query 1 TGC-AGTCGAACGGCAGCACGGAGCTTGCTCTGGTGGCGAGTGGCGAACGGGTGAGTAAT 59
||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 28 TGCAAGTCGAACGGCAGCACGGAGCTTGCTCTGGTGGCGAGTGGCGAACGGGTGAGTAAT 87
Query 60 ATATCGGAACGTACCCTGGAGTGGGGGATAACGTAGCGAAAGTTACGCTAATACCGCATA 119
||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||
Sbjct 88 ATATCGGAACGTACCCTAGAGTGGGGGATAACGTAGCGAAAGTTACGCTAATACCGCATA 147
Query 120 CGATCTAAGGATGAAAGTGGGGGATCGCAAGACCTCATGCTCGTGGAGCGGCCGATATCT 179
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 148 CGATCTAAGGATGAAAGTGGGGGATCGCAAGACCTCATGCTCGTGGAGCGGCCGATATCT 207
Query 180 GATTAGCTAGTTGGTAGGGTAAAAGCCTACCAAGGCATCGATCAGTAGCTGGTCTGAGAG 239
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 208 GATTAGCTAGTTGGTAGGGTAAAAGCCTACCAAGGCATCGATCAGTAGCTGGTCTGAGAG 267
Query 240 GACGACCAGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGG 299
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 268 GACGACCAGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGG 327
Query 300 GAATTTTGGACAATGGGCGAAAGCCTGATCCAGCAATGCCGCGTGAGTGAAGAAGGCCTT 359
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 328 GAATTTTGGACAATGGGCGAAAGCCTGATCCAGCAATGCCGCGTGAGTGAAGAAGGCCTT 387
Query 360 CGGGTTGTAAAGCTCTTTTGTCAGGGAAGAAACGGTGAGAGCTAATATCTCTTGCTAATG 419
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 388 CGGGTTGTAAAGCTCTTTTGTCAGGGAAGAAACGGTGAGAGCTAATATCTCTTGCTAATG 447
Query 420 ACGGTACCTGAAGAATAAGCACCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGG 479
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 448 ACGGTACCTGAAGAATAAGCACCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGG 507
Query 480 GTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGTGCGCAGGCGGTTTTGTAAGTCTG 539
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 508 GTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGTGCGCAGGCGGTTTTGTAAGTCTG 567
Query 540 ATGTGAAATCCCCGGGCTCAACCTGGGAATTGCATTGGAGACTGCAAGGCTAGAATCTGG 599
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 568 ATGTGAAATCCCCGGGCTCAACCTGGGAATTGCATTGGAGACTGCAAGGCTAGAATCTGG 627
Query 600 CAGAGGGGGGTAGAATTCCACGTGTAGCAGTGAAATGCGTAGATATGTGGAGGAACACCG 659
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 628 CAGAGGGGGGTAGAATTCCACGTGTAGCAGTGAAATGCGTAGATATGTGGAGGAACACCG 687
Query 660 ATGGCGAAGGCAGCCCCCTGGGTCAAGATTGACGCTCATGCACGAAAGCGTGGGGAGCAA 719
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 688 ATGGCGAAGGCAGCCCCCTGGGTCAAGATTGACGCTCATGCACGAAAGCGTGGGGAGCAA 747
Query 720 ACAGGATTAGATACCCTGGTAGTCCACGCCCTAAACGATGTCTACTAGTTGTCGGGTCTT 779
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 748 ACAGGATTAGATACCCTGGTAGTCCACGCCCTAAACGATGTCTACTAGTTGTCGGGTCTT 807
Query 780 AATTGACTTGGTAACGCAGCTAACGCGTGAAGTAGACCGCCTGGGGAGTACGGTCGCAAG 839
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 808 AATTGACTTGGTAACGCAGCTAACGCGTGAAGTAGACCGCCTGGGGAGTACGGTCGCAAG 867
Query 840 ATTAAAACTCAAAGGAATTGACGGG 864
|||||||||||||||||||||||||
Sbjct 868 ATTAAAACTCAAAGGAATTGACGGG 892
Contributed by: Matthew W. Bowman & Carly Carter, BIOL 250 – Spring 2017, Longwood University