Pseudomonas baetica is a novel fish pathogen found in the livers of flatfish wedge sole (Dicologlossa cuneata). The cells are large irregular-shaped rods that are gram-negative and motile. The colonies are small, round and whitish (López, Diéguez, Doce, Roca, Herran, Navas, Toranzo Romalde 2012). This bacteria has been isolated and cultured in Spain (López 2012)
References:
- Lopez JR, Dieguez AL, Doce A, Roca EDL, Herran RDL, Navas JI, Toranzo AE, Romalde JL. Pseudomonas baetica sp. nov., a fish pathogen isolated from wedge sole, Dicologlossa cuneata (Moreau). International Journal Of Systematic And Evolutionary Microbiology. 2012;62(Pt 4):874–882.
- López J, Navas J, Thanantong N, Herran RDL, Sparagano O. Simultaneous identification of five marine fish pathogens belonging to the genera Tenacibaculum, Vibrio, Photobacterium and Pseudomonas by reverse line blot hybridization. Aquaculture. 2012;324-325:33–38.
- Mulet M, Gomila M, Scotta C, Sánchez D, Lalucat J, García-Valdés E. Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Systematic and Applied Microbiology. 2012;35(7):455–464.
Date collected: February 2, 2017

Figure 1. Distance of site from the Appomattox River.

Figure 2. Soil collection site.

Figure 3. Colony selected for Identification.
Methods for Isolation and Identification:
- Sample was taken from the top soil of the ground right next to the Appomattox River by Lancer Park (Figure 1 and Figure 2). A sterile swab was mixed with water and speread across a sterile agar plate. The plate was then incubated at 37 degrees Celsius for 96 hours.
- A white colony (Figure 3) was selected for gene sequencing by PCR amplification
- The PCR product was digested with MspI and sequenced to identify the genus and species of the bacteria.
Results:
MspI Digestion (Figure 4): A 1,000 product was amplified by PCR. Upon digestion with MspI, two bands at roughly 200 bp and 500 bp were identified.
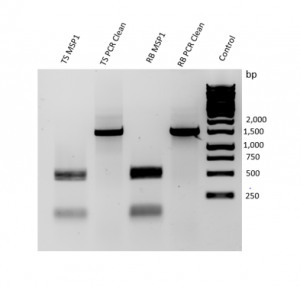
Figure 4. Results of PCR amplification and MspI Digestion.
Sequence Analysis (Figure 5): The sequenced PCR product generated 952 bases of high-quality reads that were used to identify the genus and species of the colony. NCBI BLAST analysis received 99% identity of Pseudomonas baetica.

Sequence Analysis (Figure 5)