The bacteria we found was the gram-negative bacteria Stenotrophomonas rhizophila which recently found on the roots of Brassica napus (Kai et al. 2007) and Astragalus terraccianoi and Centaurea horrida from the Mediterranean islands (Muresu et al. 2011). This bacteria is a plant root-associated bacteria that act as an anti-fungal defense for plants (Kai et al. 2007), and it is relatively harmless to humans compared to its pathogenic counterparts within the genus (Alavi P et al. 2014).
References
Alavi P, Starcher MR, Thallinger GG, Zachow C, Müller H, Berg G. 2014. Stenotrophomonas comparative genomics reveals genes and functions that differentiate benficial and pathogenic bacteria. BMC Genomics. 482(15): 1-15
Kai M, Effmert U, Berg G, Piechulla B. 2007. Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhzoctonia solani. Arch Microbiol. 187(5): 3551-360
Muresu R, Polone E, Sorbolini S, Squartini A. 2001. Characterization of endophytic and symbiotic bacteria within plants of the endemic association Centaureetum horridae Mol. Plant Biosystems. 145(2): 478-484
Date of collection
February 2, 2017
Methods for isolation and identification
- The surface soil from the banks of Buffalo Creek were sampled and collected in a 25mL collection tube (Figure 1). The sample was taken back for serial dilutions, pipette and spread with sterile hockey sticks on to LB agar plate and incubated at room temperature for 1 week.
- A medium sized light yellow colony (Figure 2) was selected for 16s rRNA gene sequencing by PCR amplification
- To identify the genus and species of the bacteria the PCR product was digested with MspI then sent off for sequencing. The BLAST was used to index the best match for the species.

Figure 1. colletion site
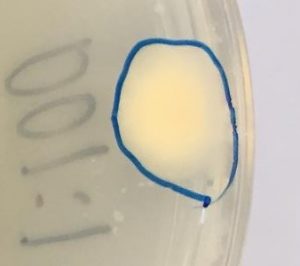
Figure 2. colony selected for identification
Results
- MspI digestion (Figure 3): The amplification of the PCR production was 1,500 bp. When digested with MspI fragments at 600 bp, 450 bp, and 200 bp were observed.
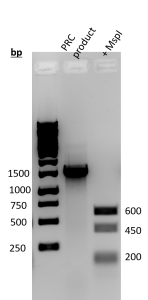
Figure 3. Result of PCR amplification and MspI digestion
- Sequence Analysis (Figure 4): 792 of high quality bases of the sequence PCR product were to index the genus and species of the selected colony. The PDF of the chromatogram can be found here 11S_PREMIX_JF7518_14. NCBI BLAST revealed that 99% of the sequence identified with 138-929 bases of the 16s rRNA gene of Strenostromonas rhizophila. (Figure 4)
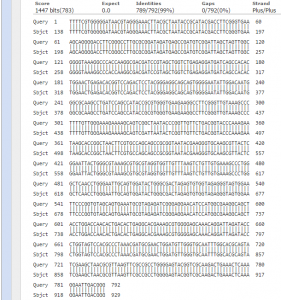
Figure 4. NCBI BLAST analysis of colony S11
Contributed by: Rex L. Liggon & Zach C. Long, BIOL 250 Spring 2017, Group 11