Pseudomonas azotoformans is an off-white, pinkish bacteria that has been found in Japanese unhulled rice (Nie, 2011). In Prince Edward County, this bacteria was collected from a Hampden-Sydney steam.
References:
Nie ZJ, Hang BJ, Cai S, Xie XT, He J, Li SP. 2011. Degradation of cyhalofop-butyl (CtB) by Pseudomonas
azotoformans strain QDZ-1 and cloning of a novel gene encoding CyB-hydrolyzing esterase. J.
Agric. Food Chem; 55(11): 6040-6.
Date Collected: January 31, 2017
Methods for Isolation and Identification:
Soil was collected from the bottom of a stream near the campus of Hampden-Sydney (Figure 1).
It was mixed with 10 mL of sterile water and it was streaked onto a nutrient-rich agar plate using the
hockey method. An off-white, pinkish bacterial colony was chosen to have its DNA sequenced by PCR amplification and identified through the BLAST database on NCBI (Figure 2).
After the PCR was produced, a restriction enzyme, MSP 1, was used to digest the DNA and it was run through a gel. It was sent off for sequencing to identify the bacteria.

Figure 1. Sample collection site
 Figure 2. Colony selected for identification.
Figure 2. Colony selected for identification.
Results:
- MSP1 digestion (Figure 3): A 2,000 bp product was amplified through the PCR method. One band was identified at approximately 250 bp after it was digested with MSP1.
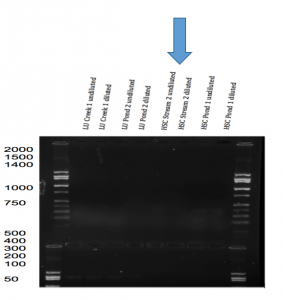
Figure 3. Result of MSP 1 digestion and PCR amplification test through Gel Electrophoresis
- Sequence Analysis (Figure 4): 709 bases were produced in the PCR product to be analyzed. With this product, clear nucleotide base sequences were used from SnapGene Viewer to be used as the sequence for a targeted loci BLAST in the ncbi BLAST database. The chromatogram can be viewed: (TGCEHS2_PREMIX_JF7573_41). The BLAST data revealed that a 99% identity between the query and the potential microbe from base pairs 29-736. Within those base pairs, there were only 2 gaps between the two. The bacteria was identified as Pseudomonas azotoformans (Figure 4).
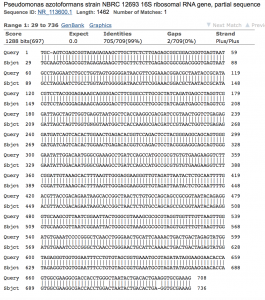
Figure 4. NCBI Blast analysis of Pseudomonas azotoformans strain NBRC
Contributed by: Tatianna Griffin and Cecilie Elliott, BIOL 250-50 Spring 2017