The bacteria we found, Pseudomonas Vancouvernsis, is a gram- negative soil bacterium that grows on pulp mill effluents with resin acids. This bacteria has been discovered in the North Western Indian Himalayas where it shows a wide pH range (5–12; optimum 7.0) and salt concentrations up to 5% (w/v) (Mishra, 2008).
References
Methods for isolation and Identification:
- There were two particular surfaces that were collected and tested from the Buffalo creek near the Environmental education center ( Figure 1). While collecting the water we tested 100μl of water, 100μl of 1:10 dilution of water, and 100μl of 1:100 dilution of water. For the soil, 0.5 grams of soil was added to 50 ml of sterile water and the same amounts were tested.
- A small colony from our undiluted water plate and a big colony from our 1:100 soil plate were selected for 16S rRNA gene sequencing by PCR amplification ( Figure 2).
- The product we received was digested and mixed with Msp1; later the product was sent to a sequencing center.Once we received them back, we were able to specify the genus and species of our bacteria.

Figure 1. Collection
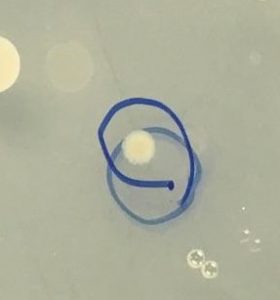
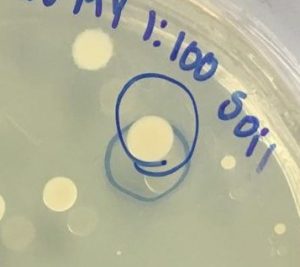
Figure 2. Colony selected for identification
Results:
- Msp1 digestion (figure 3.): A 1,550 bp product was amplified by PCR. With the digestion of Msp1, there really aren’t any bands that are noticeable until it gets closer to the base pair lane.
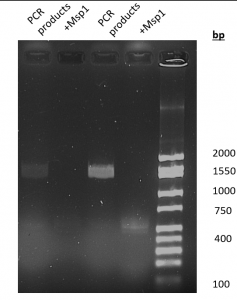
Figure 3. Results of PCR amplification and MspI digestion
- Sequence analysis (figure 4.): The sequenced PCR product generated 812 bases of high-quality reads that were used to identify the genus and species of the colony. The chromatogram of the sequence is available as a pdf (BMB_PREMIX_JF7535_7). NCBI BLAST analysis showed 96% identity with bases 28-838 of the 16s rRNA gene of Psuedomonas vancouverensis (Figure 4).
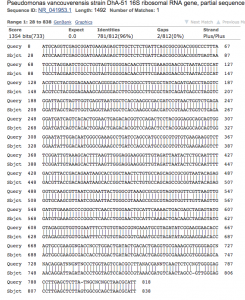
Figure 4. NCBI BLAST analysis of colony 16-1
Contributed by: Maya Young and Briana Smith, BIOL 250 Spring 2017