Pseudomonas trivialis is a fluorescent, Gram-negative bacterium isolated from the phyllosphere of grasses. This bacterium has been isolated from the rhizosphere of Hippophae rhamnoides growing in the Lahaul-Spiti district, India.
Citation:
Mejri D, Gamalero, E, Souissi T. 2012. Formulation development of the deleterious rhizobacterium Pseudomonas trivialis X33d for biocontrol of brome (Bromus diandrus) in durum wheat. Journal of Applied Microbiology. 114:2019-228.
Date Collected: February 8, 2017
Methods for Isolation & Identification:
- Collection and Sampling of Environmental Samples at Environmental Education Center
- For each collection site, we labeled 3 nutrient agar plates as follows: “direct count,” “1:10,” & “1:100“
- To plate these samples, we used the serial dilution technique
- Analyzing Data
- After 7 days, we collected the following data for each plate: number of colonies, color, size, shape, texture, form, elevation, and margin.
- Genomic DNA Isolation/Extraction
- Genomic DNA was extracted from two colonies to identify the bacterial species.
- Polymerase Chain Reaction (PCR)
- The 16s rDNA sequence of our own two unknown bacteria was amplified.
- Restriction Enzyme Digestion & Gel Electrophoresis
- The purpose of this step was to cleave/ligate a functional piece of DNA predictably and precisely (Restriction Enzyme Digestion).
- After cleavage, DNA fragments was separated using agarose gel electrophoresis.
- These steps will result in the DNA sequencing step.
- DNA Sequencing Analysis/Identification
- The purpose of this step was to identify our prokaryotes based on the sequence of the 16s rDNA. Also, analyzing our DNA sequences and identifying what prokaryote we likely identified.
- SnapGene viewer to analyze our sequence, used a BLAST to identify the prokaryote, and checked the MspI digestion sites for our sequence using NEB cutter
Results:
The sequenced PCR product for the 16s rRNA gene generated 857 bases of high-quality reads that were used to identify the genus and species of the bacteria colonies. This allowed us to separate and analyze the DNA and their fragments, based on their size and charge. The Gel Electrophoresis image below shows the nucleic acid molecules separated by bands. The shorter molecules moved faster and traveled farther than the longer ones because the shorter molecules traveled through the pores of the gel. As you can clearly see in the image below bands lining up, which represents a protein that compromises that band is highly abundant.
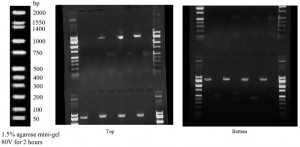
The digested gels are 3, 5, 7, and 9 with the top being Buffalo Creek tree at 5m, tree at 10m, and the two water samples.
Sequence Analysis:
The sequenced PCR product generated 865 bases of high-quality reads that were used to identify the genus and species of the colony. The chromatogram of the sequence is available as a pdf (BCW2) . NCBI BLAST analysis revealed 99% identity with bases 29-988 of the 16s rRNA gene of Pseudomonas trivialis.
| Score | Expect | Identities | Gaps | Strand |
|---|---|---|---|---|
| 1735 bits(939) | 0.0 | 954/961(99%) | 1/961(0%) | Plus/Plus |
Query 1 TGCCAGTCGAGCGGTAGAGAGGAGCTTGCTTCTCTTGAGAGCGGCGGACGGGTGAGTAAT 60
||| ||||||||||||||||| ||||||||||||||||||||||||||||||||||||||
Sbjct 29 TGCAAGTCGAGCGGTAGAGAGAAGCTTGCTTCTCTTGAGAGCGGCGGACGGGTGAGTAAT 88
Query 61 GCCTAGGAATCTGCCTGGTAGTGGGGGATAACGTTCGGAAACGGACGCTAATACCGCATA 120
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 89 GCCTAGGAATCTGCCTGGTAGTGGGGGATAACGTTCGGAAACGGACGCTAATACCGCATA 148
Query 121 CGTCCTACGGGAGAAAGCAGGGGACCTTCGGGCCTTGCGCTATCAGATGAGCCTAGGTCG 180
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 149 CGTCCTACGGGAGAAAGCAGGGGACCTTCGGGCCTTGCGCTATCAGATGAGCCTAGGTCG 208
Query 181 GATTAGCTAGTTGGTGAGGTAATGGCTCACCAAGGCGACGATCCGTAACTGGTCTGAGAG 240
|||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||
Sbjct 209 GATTAGCTAGTTGGTGGGGTAATGGCTCACCAAGGCGACGATCCGTAACTGGTCTGAGAG 268
Query 241 GATGATCAGTCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGG 300
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 269 GATGATCAGTCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGG 328
Query 301 GAATATTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTT 360
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 329 GAATATTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTT 388
Query 361 CGGATTGTAAAGCACTTTAAGTTGGGAGGAAGGGCAGTTACCTAATACGTGATTGTTTTG 420
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 389 CGGATTGTAAAGCACTTTAAGTTGGGAGGAAGGGCAGTTACCTAATACGTGATTGTTTTG 448
Query 421 ACGTTACCGACAGAATAAGCACCGGCTAACTCTGTGCCAGCAGCCGCGGTAATACAGAGG 480
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 449 ACGTTACCGACAGAATAAGCACCGGCTAACTCTGTGCCAGCAGCCGCGGTAATACAGAGG 508
Query 481 GTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTAGGTGGTTTGTTAAGTTGG 540
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 509 GTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTAGGTGGTTTGTTAAGTTGG 568
Query 541 ATGTGAAATCCCCGGGCTCAACCTGGGAACTGCATTCAAAACTGACTGACTAGAGTATGG 600
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 569 ATGTGAAATCCCCGGGCTCAACCTGGGAACTGCATTCAAAACTGACTGACTAGAGTATGG 628
Query 601 TAGAGGGTGGTGGAATTTCCTGTGTAGCGGTGAAATGCGTAGATATAGGAAGGAACACCA 660
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 629 TAGAGGGTGGTGGAATTTCCTGTGTAGCGGTGAAATGCGTAGATATAGGAAGGAACACCA 688
Query 661 GTGGCGAAGGCGACCACCTGGACTAATACTGACACTGAGGTGCGAAAGCGTGGGGAGCAA 720
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 689 GTGGCGAAGGCGACCACCTGGACTAATACTGACACTGAGGTGCGAAAGCGTGGGGAGCAA 748
Query 721 ACAGGATTAGATACCCTGGTAGTCCACGCCGTAAACGATGTCAACTAGCCGTTGGAAGCC 780
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 749 ACAGGATTAGATACCCTGGTAGTCCACGCCGTAAACGATGTCAACTAGCCGTTGGAAGCC 808
Query 781 TTGAGCTTTTAGTGGCGCAGCTAACGCATTAAGTTGACCGCCTGGGGAGTACGGCCGCAA 840
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 809 TTGAGCTTTTAGTGGCGCAGCTAACGCATTAAGTTGACCGCCTGGGGAGTACGGCCGCAA 868
Query 841 GGTTAAAACTCAAATGAATTGACGGGGGCCCGCACAAGCGGTGGAGCATGTGGTTTAATT 900
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 869 GGTTAAAACTCAAATGAATTGACGGGGGCCCGCACAAGCGGTGGAGCATGTGGTTTAATT 928
Query 901 CGAAGCAACGCGAAAGAACCTTACCAGGGCTTTGCATCCAATGAACTTTCTAGAGATAGA 960
|||||||||||| ||||||||||||||| ||| ||||||||||||||||||||||||||
Sbjct 929 CGAAGCAACGCG-AAGAACCTTACCAGGCCTTGACATCCAATGAACTTTCTAGAGATAGA 987
Query 961 T 961
|
Sbjct 988 T 988
Contributed by: Matthew W. Bowman & Carly Carter, BIOL 250 – Spring 2017, Longwood University