Pseudomonas extremoustralis is a Gram-negative and rod-shaped bacterium. This bacterium is commonly found in colder environments. This bacterium as been isolated from a temporary pond in Antarctica/cold environments.
Citation:
Tribelli PM, Venero ECS, Ricardi MM, Gomez-Lozano M, Raiger lustman LJ, Molin S, Lopez NI. 2015. Novel essential role of ethanol oxidation genes at low temperature revealed by transciptome analysis in the Antarctic bacterium Pseudomonas extremaustralis. PLoS One. 10(12)
Data Collected: February 8, 2017
Methods for Isolation & Identification:
- Collection and Sampling of Environmental Samples at Environmental Education Center
- For each collection site, we labeled 3 nutrient agar plates as follows: “direct count,” “1:10,” & “1:100“
- To plate these samples, we used the serial dilution technique
- Analyzing Data
- After 7 days, we collected the following data for each plate: number of colonies, color, size, shape, texture, form, elevation, and margin.
- Genomic DNA Isolation/Extraction
- Genomic DNA was extracted from two colonies to identify the bacterial species.
- Polymerase Chain Reaction (PCR)
- The 16s rDNA sequence of our own two unknown bacteria was amplified.
- Restriction Enzyme Digestion & Gel Electrophoresis
- The purpose of this step was to cleave/ligate a functional piece of DNA predictably and precisely (Restriction Enzyme Digestion).
- After cleavage, DNA fragments was separated using agarose gel electrophoresis.
- These steps will result in the DNA sequencing step.
- DNA Sequencing Analysis/Identification
- The purpose of this step was to identify our prokaryotes based on the sequence of the 16s rDNA. Also, analyzing our DNA sequences and identifying what prokaryote we likely identified.
- SnapGene viewer to analyze our sequence, used a BLAST to identify the prokaryote, and checked the MspI digestion sites for our sequence using NEB cutter
Results:
The sequenced PCR product for the 16s rRNA gene generated 857 bases of high-quality reads that were used to identify the genus and species of the bacteria colonies. This allowed us to separate and analyze the DNA and their fragments, based on their size and charge. The Gel Electrophoresis image below shows the nucleic acid molecules separated by bands. The shorter molecules moved faster and traveled farther than the longer ones because the shorter molecules traveled through the pores of the gel. As you can clearly see in the image below bands lining up, which represents a protein that compromises that band is highly abundant.
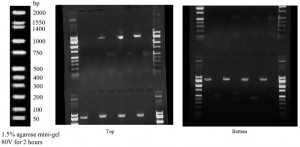
The digested gels are 3, 5, 7, and 9 with the top being Buffalo Creek tree at 5m, tree at 10m, and the two water samples.
Sequence Analysis:
The sequenced PCR product generated 865 bases of high-quality reads that were used to identify the genus and species of the colony. The chromatogram of the sequence is available as a pdf (BCT10) . NCBI BLAST analysis revealed 99% identity with bases 57-960 of the 16s rRNA gene of Pseudomonas extremaustralis.
Pseudomonas extremaustralis strain 14-3 16S ribosomal RNA, complete sequence
| Score | Expect | Identities | Gaps | Strand |
|---|---|---|---|---|
| 1626 bits(880) | 0.0 | 897/905(99%) | 2/905(0%) | Plus/Plus |
Query 1 GTCGAGCGGTAGAGAGAAGCTTGCTTCTCTTGAGAGCGGCGGACGGGTGAGTAATGCCTA 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 57 GTCGAGCGGTAGAGAGAAGCTTGCTTCTCTTGAGAGCGGCGGACGGGTGAGTAATGCCTA 116
Query 61 GGAATCTGCCTGGTAGTGGGGGATAACGCTCGGAAACGGACGCTAATACCGCATACGTCC 120
|||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||
Sbjct 117 GGAATCTGCCTGGTAGTGGGGGATAACGTTCGGAAACGGACGCTAATACCGCATACGTCC 176
Query 121 TACGGGAGAAAGCAGGGGACCTTCGGGCCTTGCGCTATCAGATGAGCCTAGGTCGGATTA 180
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 177 TACGGGAGAAAGCAGGGGACCTTCGGGCCTTGCGCTATCAGATGAGCCTAGGTCGGATTA 236
Query 181 GCTAGTTGGTGAGGTAATGGCTCACCAAGGCGACGATCCGTAACTGGTCTGAGAGGATGA 240
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 237 GCTAGTTGGTGAGGTAATGGCTCACCAAGGCGACGATCCGTAACTGGTCTGAGAGGATGA 296
Query 241 TCAGTCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGGGAATA 300
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 297 TCAGTCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGGGAATA 356
Query 301 TTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTTCGGAT 360
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 357 TTGGACAATGGGCGAAAGCCTGATCCAGCCATGCCGCGTGTGTGAAGAAGGTCTTCGGAT 416
Query 361 TGTAAAGCACTTTAAGTTGGGAGGAAGGGCAGTTACCTAATACGTGATTGTTTTGACGTT 420
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 417 TGTAAAGCACTTTAAGTTGGGAGGAAGGGCAGTTACCTAATACGTGATTGTTTTGACGTT 476
Query 421 ACCGACAGAATAAGCACCGGCTAACTCTGTGCCAGCAGCCGCGGTAATACAGAGGGTGCA 480
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 477 ACCGACAGAATAAGCACCGGCTAACTCTGTGCCAGCAGCCGCGGTAATACAGAGGGTGCA 536
Query 481 AGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTAGGTGGTTCGTTAAGTTGGATGTG 540
|||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||
Sbjct 537 AGCGTTAATCGGAATTACTGGGCGTAAAGCGCGCGTAGGTGGTTTGTTAAGTTGGATGTG 596
Query 541 AAATCCCCGGGCTCAACCTGGGAACTGCATTCAAAACTGTC-GAGCTAGAGTATGGTAGA 599
||||||||||||||||||||||||||||||||||||||| | || |||||||||||||||
Sbjct 597 AAATCCCCGGGCTCAACCTGGGAACTGCATTCAAAACTGACTGA-CTAGAGTATGGTAGA 655
Query 600 GGGTGGTGGAATTTCCTGTGTAGCGGTGAAATGCGTAGATATAGGAAGGAACACCAGTGG 659
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 656 GGGTGGTGGAATTTCCTGTGTAGCGGTGAAATGCGTAGATATAGGAAGGAACACCAGTGG 715
Query 660 CGAAGGCGACCACCTGGACTGATACTGACACTGAGGTGCGAAAGCGTGGGGAGCAAACAG 719
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 716 CGAAGGCGACCACCTGGACTGATACTGACACTGAGGTGCGAAAGCGTGGGGAGCAAACAG 775
Query 720 GATTAGATACCCTGGTAGTCCACGCCGTAGACGATGTCAACTAGCCGTTGGGAGCCTTGA 779
||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||
Sbjct 776 GATTAGATACCCTGGTAGTCCACGCCGTAAACGATGTCAACTAGCCGTTGGGAGCCTTGA 835
Query 780 GCTCTTAGTGGCGCAGCTAACGCATTAAGTTGACCGCCTGGGGAGTACGGCCGCAAGGTT 839
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 836 GCTCTTAGTGGCGCAGCTAACGCATTAAGTTGACCGCCTGGGGAGTACGGCCGCAAGGTT 895
Query 840 AAAACTCAAATGAATTGACGGGGGCCCGCACAAGCGGTGGAGCATGGGGTTTAATTCCAA 899
|||||||||||||||||||||||||||||||||||||||||||||| |||||||||| ||
Sbjct 896 AAAACTCAAATGAATTGACGGGGGCCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGAA 955
Query 900 GCAAC 904
|||||
Sbjct 956 GCAAC 960
Contributed by: Matthew W. Bowman & Carly Carter, BIOL 250 – Spring 2017, Longwood University