Flavobacterium sacchorophilum is a Gram-negative, nonmotile and motile, rod-shaped bacteria that was first isolated on the surface of the Gulkana Glacier in 2001. It can also be found in the Buffalo Creek in Prince Edward County.
Reference:
- Segawa, Takahiro, Yoshitaka Yoshimura, Kenichi Watanabe, Hiroshi Kanda, and Shiro Kohshima. “Community structure of culturable bacteria on surface of Gulkana Glacier, Alaska.”Polar Science 1 (2011): 41-51. Web.
Date collected: February 8, 2017
Methods for isolation and identification:
- Collection and Sampling of Environmental Samples at Environmental Education Center (Figure 1)
- Selection of bacteria from plate to be analyzed (Figure 2)
- Polymerase Chain Reaction to amplify 16S rRNA
- Restriction Enzyme Digestion & Gel Electrophoresis (Figure 3)
- DNA Sequencing Analysis/Identification

Figure 1. Sampling site locations in Lancer Park. The top site is Buffalo Creek and the lower site is the pond where samples were taken
Figure 2: picture of Flavobacterium sacchorophilum
Results:
- MspI digestion (Figure 3): A 1,500 bp product was amplified by PCR. Upon digestion with a band at 500 bp digested and a 1000 bp undigested.
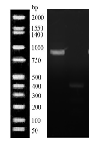
Figure 3: Mspi digestion of Flavobacterium sacchorophilum PCR product
- Sequence analysis (Figure 4): The sequenced PCR product generated 994 bases. NCBI BLAST analysis revealed 99% identity with 993 of the bases matching the 16s rRNA gene of Flavobacterium sacchorophilum (Figure 4).
 Figure 4: The BLAST sequence match of the sample bacteria to Flavobacterium sacchorophilum.
Figure 4: The BLAST sequence match of the sample bacteria to Flavobacterium sacchorophilum.
Contributed by: Matthew W. Bowman & Carly Carter, BIOL 250 – Spring 2017, Longwood University
Methods for Isolation and Identification:
- A soil sample was taken from the bottom of the Buffalo Creek (Figure 1). 100μl of the sample was placed on an agar plate and was incubated at 25 degrees Celsius for 48 hours. The plate was parafilmed and placed in the refrigerator for seven days.
- A light orange-ish/whitish, circular colony (Figure 2) was selected for 16S rRNA gene sequencing by PCR amplification.
- The PCR product was sent for DNA sequencing to identify the species of the bacteria.

Figure 1. site of collection (Buffalo Creek)

Figure 2. Selected colony for identification
Results:
- Msp1 digestion (figure 3): A bold band was produced at 750 bp by PCR.
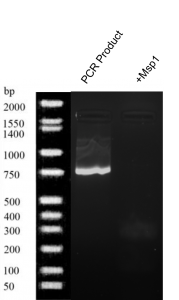
Figure 3. Gel electrophoresis results of Buffalo Creek soil PCR product and + MSP1.
- DNA sequence: The sequenced PCR product produced 875 quality base pairs that were used to identify the bacteria as Flavobacterium sacchorophilum. The chromatogram of the sequence is available as a pdf (HMX1_PREMIX_JF7558_27). NCBI BLAST revealed 98% similarity with 2 gaps out of 875 base pairs (Figure 4).

Figure 4. NCBI BLAST results of chosen colony.
Contributed by: Megan Bland and Hannah Hatke, BIOL 250 Spring 2017