BES Faculty-Student Research
Students interested in conducting research with faculty in fall 2024 should fill out this form.
Dale Beach, Ph.D.
Betting on Biotechnology to Build Better Brewers
*** Dr. Beach is always willing to work with curious students interested in Cellular and Molecular Biology. Please email to set up an appointment. ***
The common yeast, Saccharomyces cerevisiae, is frequently associated with baking and beer brewing where t he yeast consumes sugars to produce carbon dioxide gas – important as a rising agent in bread dough – and ethanol – the inebriating factor in beer, wine, and distilled spirits. On an industrial scale, baking and alcohol production (for human consumption and for commercial uses), represents billions of dollars annually. As you might guess, the commercial importance of yeast-based fermentation has given rise to a vast catalog of different yeast varieties that a fermenter, baker, or brewer can use to optimize their product quality, increase productivity, and lower costs. Surprisingly, the genetics of this industrially important organism is poorly understood from an industrial point of view. In the Beach lab we are working to understand the genetic roots of some very complicated yeast traits so that we can rapidly develop new strains for the craft brewing industry. By identifying potential target genes in the yeast genome, we are using DNA sequence analysis to characterize variations in gene sequences to better correlate genes with ideal traits, and the use that knowledge to generate novel yeast strains for the fermentation industry. We are currently investigating genes in the Flocculation pathway, an important process to collect and remove yeast cells from the fermentation reaction without costly filtration equipment. Working on this project will help you develop DNA manipulation and molecular biology skills as well as computational analysis of genomic DNA sequences. Email for more information: beachdl@longwood.edu.

Adam Franssen, Ph.D.
Behavioral Neuroscience in Rats
*** Dr. Franssen is seeking one new research student for fall 2024. ***
In broad terms, the Franssen lab is investigating how the developmental environment affects gene expression, brain chemistry, animal behavior, and ultimately reproductive success in animals. Specifically, students currently working with rats in our state-of-the-art Neuroscience Facility in Allen Hall are investigating how two factors – maternal behavior and environmental enrichment – affect the cognitive abilities of offspring. Students working in the lab will learn the knowledge and skill needed to conduct behavioral neuroscience research. Skills may include animal husbandry, behavioral testing, analysis of behavior, cryosectioning of neural tissue, immunohistochemistry, neuroquantification, written and oral communication. Once those skills are mastered, each undergraduate research collaborator is encouraged to develop their own project to work on. For example, one recent student-motivated research program has investigated the epigenetically heritable consequences have having a “Good” or “Bad” mother. Another research projects seeks to determine whether environmental enrichment or loss during early development can alter behavioral responses to new stimuli.
Students wishing to join the lab should be highly motivated, be able to work independently after training, and be excited to collaborate with others from different backgrounds. Students from all majors/minors are welcome; successful members of the Franssen Lab have come from Biology, Chemistry, Kinesiology, Psychology, and Neuroscience Studies programs. Varied career goals are welcome, too. In addition to multiple Neuroscience PhDs, Franssen Lab Alumni have become Anesthesiologists, Dietitians, Genetic Counselors, Lawyers, Physical Therapists, and Teachers.

Jameson Hinkle, M.S.
Demographic and genetic (Demogenetic) research simulations for reintroductions of species
*** Mr. Hinkle is seeking one new research student for fall 2024. ***
Research into demogenetic outcomes of reintroductions of species is critical to maintaining biodiversity in the face of climate change. Previous work has indicated that we can best understand where to reintroduce a threatened Char species into a currently extirpated watershed in order to help maintain the natural ecosystem. In this research, students will participate in understanding how simulations in Python, and analysis in R and GIS can help policy makers make decisions around how to maintain fisheries in the face of climate change through understanding demogenetic changes in space and time.
Brandon Jackson, Ph.D.
*** Dr. Jackson will be accepting one new research student for fall 2024. ***
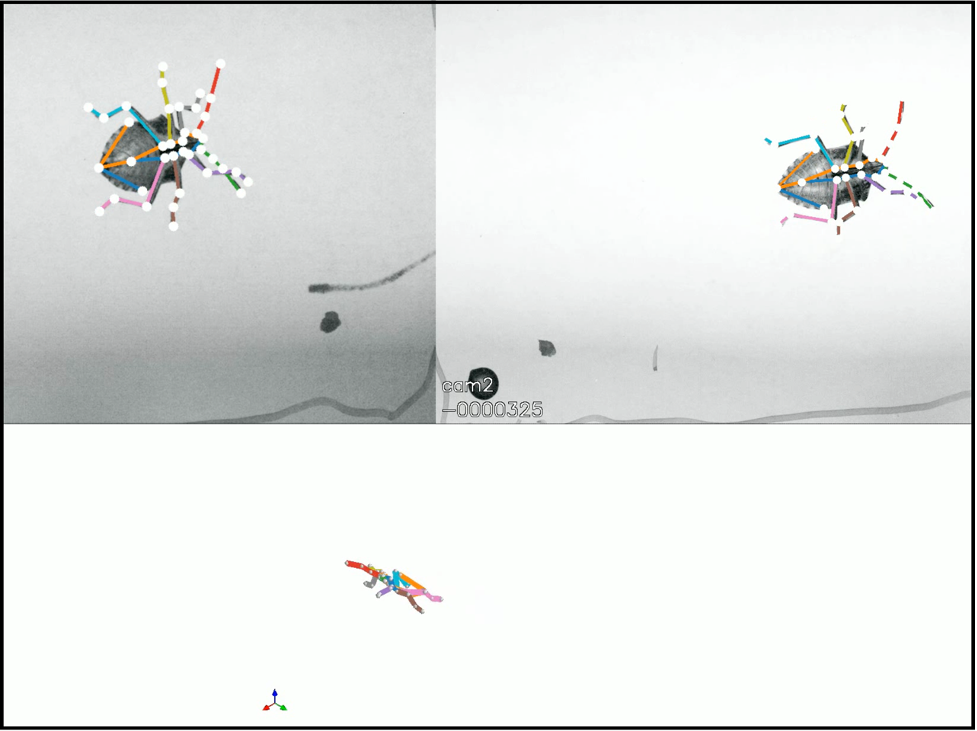
How do animals move? When different animals have differently shaped legs, wings, or bodies, does that affect how well they move? My lab studies the evolution of anatomy and physiology related to animal locomotion, both in the lab and in the field. We ask questions related to neuroscience, animal behavior, developmental biology, ecology, biomechanics, physiology, and evolution, but students do not need a background in any of the above to start in the lab.
Research students are encouraged to develop their own projects based on their interests but can also work on existing projects. We study nearly any type of animal. I currently have active projects studying the following: how losing a leg affects walking in stinkbugs; unique properties of ladybug walking and takeoff; the role of bird tails in slow-speed flight; the role of wing flexibility in cicada flight; crawfish fighting mechanics; and startled zebrafish swimming.
Example videos can be found at my lab page: https://blogs.longwood.edu/jacksonlab/
Björn Ch. Ludwar, Ph.D.
Dermatoglyphics using Neuronal Networks and Bioimaging Lab
*** Dr. Ludwar is seeking two new research students for spring 2024 ***
I am looking for self-motivated, curious students, interested in working in relative independence on one of the following projects:
1.) Dermatoglyphics – the scientific study of fingerprints – is widely used in criminology. However, it is also useful for diagnostic screening of various health conditions, such as e.g. diabetes. Recent advances in machine learning have opened up a novel approach for such screening methods. I am seeking a highly motivated student who is interested in working with me on the development of neural network based machine learning algorithms to analyze fingerprint symmetry. The project will require some knowledge of Python, TensorFlow, and Keras – or the willingness/time investment to learn.
2.) The Bioimaging Lab at Longwood University uses various imaging techniques to study biology. Current techniques include high speed videography, photogrammetry, fluorescence microscopy, and electron scanning microscopy (SEM). Projects are done in collaboration with other faculty members, or as stand-alone research projects. Students are welcome bring their own electron microscopy project. Over the course of a semester they will learn how to prepare the sample (fixation, dehydration, contrast enhancement, etc.), how to use a state-of-the-art Hitachi TM4000Plus II microscope, and how to image their samples.


Denis Trubitsyn, Ph.D.
*** Dr. Trubitsyn is always accepting research students and welcomes your inquiry. ***
Magnetic bacteria
My lab studies a unique group of bacteria known as magnetotactic. These organisms synthesize their own magnets (see attached image) and use them similarly to a compass needle in navigation. In addition to learning molecular mechanisms involved in the process of making magnets, our research focusses on application of bacterial nanoparticles in medicine. Specifically, we aim to add proteins to magnetic crystals that will make them bind to human cancer cells. Such modified magnets can then be used to aid in tumor localization with MRI.
While the projects outlined above may require several years of lab work to complete, students in my lab help to accomplish individual phases of this research. They gain skills such as preparation of culture media, growth and harvesting of various bacteria, PCR, cloning, DNA and protein gel electrophoresis, isolation of bacterial magnetic particles, detection of functional proteins with various molecular biology techniques. Students also get to present their work at state and national conferences. There are no formal prerequisites to join my lab; however, I look for someone who is enthusiastic and curious. It may be possible to develop additional research projects based on your personal interest in microbiology.

Amorette Barber, Ph.D.
*** Dr. Barber is not currently accepting research students. ***
Tumor Immunology Research Lab
My research lab focuses on enhancing immune responses to cancer. Current cancer treatments such as surgery, chemotherapy, and radiation result in adverse side effects. Therefore, the development of novel therapies that specifically target tumor cells and minimize damage to healthy cells is desirable. One option is to use cells of the immune system, specifically T cells, which kill cells that appear dangerous or foreign. To maximize tumor targeting by T cells, genetic engineering is used to express receptors that enhance tumor cell recognition. These receptors, named chimeric antigen receptors (CARs), endow the T cell with a way to recognize the tumor cells and activate many cellular functions to eradicate the tumor. Encouragingly, CAR-expressing T cells have recently received FDA-approval for cancer therapy and the chimeric antigen receptor that I developed at Longwood will be starting Phase I clinical trials for cancer therapy soon. My current research at Longwood University focuses on studying how to enhance T cell immunotherapy for many different types of cancer through 1) creation and testing of novel CARs, 2) investigation of immune cell function, and 3) study of how various compounds (including natural products and parabens) alter immune cell function. In addition to the applications to human health, my research also has implications in enhancing our understanding of general immunology and cancer therapies.

Larry Collins, Ph.D.
Coastal Resilience, Climate Change Education Research
*** Dr. Collins is not currently accepting research students. ***
Research in my lab investigates human perception of climate change risk and explores the emerging relationships between global climate change and human health. I am seeking one research student to work on the project described below for Spring 2024.
Extreme Heat and Climate Change Risk Perception– The U.S. Global Climate Change Research Program has identified climate change as a growing threat to human health. This project seeks to understand the factors that contribute and interact to influence climate change risk perceptions and professional action on climate change in central Virginia through a focus on extreme heat. This will entail working with students to design a survey that will measure the relationships between individuals’ conceptions of extreme heat and their perception of climate change risk. In this project, we also hope to learn more about how extreme heat and exposure to wildfire smoke have shaped individuals’ understanding of the relationships between climate change and human health.
Ken Fortino, Ph.D.
*** Dr. Fortino is not currently accepting research students. ***
Small, human–constructed ponds are a dominant landscape feature in eastern North America, where they can have significant impacts on watershed hydrology, biodiversity, and organic matter processing. My lab studies how small ponds alter the way that watersheds process energy and organic matter. My students and I have investigated the way ponds affect the decomposition and storage of terrestrial leaf litter and have found that leaves break down slowly when they enter ponds, indicating that ponds contribute to organic matter storage in watersheds. We have also found that inorganic nutrients are abundant in these small ponds, supporting high levels of primary productivity which interacts with leaf decomposition in a process called priming. Overall the student researchers in my lab have contributed to our understanding of ponds as important and dynamic biological systems in watersheds. My lab is not currently accepting new students for the 2023–2024 academic year but I am always happy to answer questions and explore future student research opportunities.

Mark Fink, Ph.D.
*** Dr. Fink is not currently accepting research students. ***
Longwood Campus Breeding Bird Atlas Project
The overall goal of this project is to initiate a long-term monitoring plan for avian populations on Longwood campuses (Main campus, Lancer Park, and Baliles Center at Hull Springs). This project will launch in spring 2023 (mid-January through mid-June) with breeding bird surveys of main campus and Lancer Park. Student researchers will spend the early part of the semester researching avian census methods and developing protocols for our breeding bird atlas project. Researchers will then implement these protocols by collecting field data of breeding birds on main campus and Lancer Park from March through mid-June. Compilation and analysis of data will occur concurrently with data collection. Opportunities for continuing research in subsequent semesters are possible. Students must be available through mid-June and are responsible for their own housing. Previous experience is not required though ability to identify common birds by sight and sound is preferred. Email for more information: finkml@longwood.edu.
Dina Leech, Ph.D.
*** Dr. Leech is not currently accepting research students. ***
Water Quality and Ecosystem Restoration
Land use and land cover are closely linked to coastal water quality. Runoff of nutrients and organic matter from agricultural fields often leads to algal blooms and the formation of ‘dead zones’, or regions of low dissolved oxygen, that negatively affect aquatic life. At Longwood’s Baliles Center for Environmental Education at Hull Springs, within the Chesapeake Bay Watershed, agricultural fields have recently been restored to forest and wetland. The Leech Lab is monitoring changes in local water quality on and near the property in consort with these restoration efforts to assess their effectiveness.
Students get hands-on experience collecting water samples in the field and then processing them in the lab to measure the amount of bacteria, algae, nutrients, and organic matter. We also maintain a water quality sonde for the Longwood Environmental Observatory (LEO) that collects data every 15 minutes on several water quality parameters, including water temperature, dissolved oxygen, pH, and salinity. These important monitoring data are shared publicly to better inform management strategies within the Bay’s watershed. They also complement experimental work conducted by the lab to better understand mechanisms of ‘dead zone’ formation.

Mary Lehman, Ph.D.
*** Dr. Lehman is not currently seeking new research students ***
Allelopathy and Interactions of Plant Stresses
Allelopathy involves the interaction of plants through their release of chemicals into the soil, often negatively affecting the growth of surrounding plants. This can be thought of as “chemical warfare” among plants. There are also many other stresses that impact plants, such as low nutrient availability, high temperature, and the presence of pollutants. Little is known about how other plant stresses interact with allelopathy, or how various allelochemicals interact with each other. My lab uses a nutrient culture system (hydroponics) to grow bioassay plant seedlings to precisely control exposure of plants to allelochemicals and investigates the interaction of this stress with other plant stresses.
No experience is required, but students should be prepared for the fact that this research involves the preparation of many chemical solutions; it’s not just growing plants. Students also must be committed to daily maintenance of plants while an experiment is in progress, including weekends.

Ann Mayo, Ph.D.
*** Dr. Mayo is not currently accepting research students, but is willing to discuss research interests. ***
Insect Behavior, Diversity, and Ecology
Ants, though small and often inconspicuous members of ecosystems, have enormous impacts on ecosystem health and resilience. Because most ants are ground nesting, they make a large contribution on soil generation. They are also major predators and important prey. Their species diversity and abundance tells us a lot about ecosystem health, resilience, and function. There are also important invasive ant species that upset these roles and in turn impact ecosystem health. With concerns for the impacts of pollution, human land use changes, and climate change, monitoring ant diversity has become an important tool for reclamation and conservation projects as well as land management procedures.
I am currently identifying and monitoring local ant populations (in and near Longwood’s campus) and at the Baliles Center located on the Potomac River with the aim to describe the current ant diversity and ecosystem.
Ravi Sankar, Ph.D.
*** Dr. Sankar is not currently seeking research students. ***
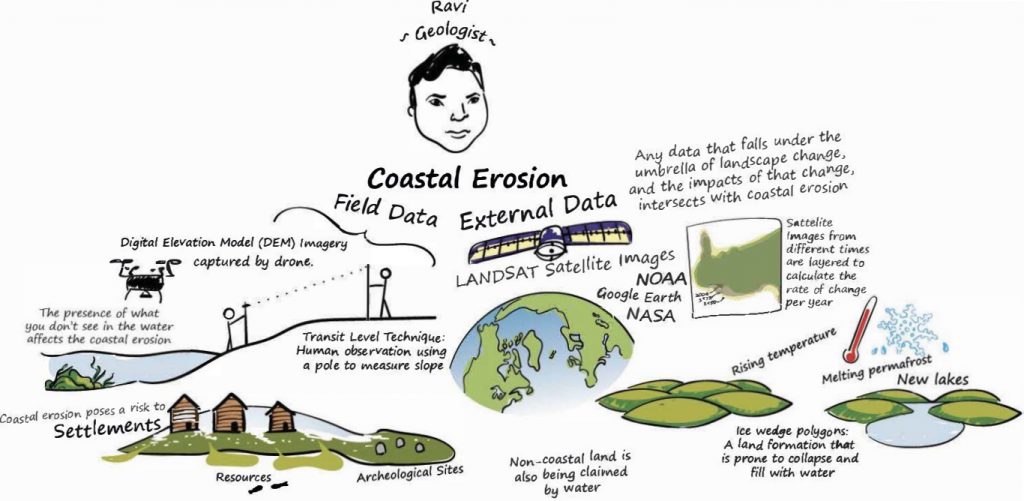
The Geomorphology Research Lab (GRL) at Longwood University employs advanced techniques such as high-resolution geospatial, topographical, and sedimentary methods to investigate how natural processes and human activities shape Earth’s landscapes over time. Improving the accuracy of datasets and computational models used to predict the frequency and severity of extreme weather events is crucial, as it highlights the significance of addressing climate variability’s short-term forecasts that indicate a potential rise in environmental risks. At the GRL, we employ various techniques (e.g. Unmanned Aerial Vehicles (UAVs), Ground-Penetrating Radar (GPR), and topographic surveys) to identify, track, and study changes resulting from sea level rise, storms, and land-use along varying coastal and fluvial regions such as beaches, bays, barrier islands, dunes, rivers, and the nearshore. These collected data are then utilized in numerical models and GIS-based techniques to investigate the intricate processes responsible for shaping the earth’s landscapes. The overall goal of our research at the GRL is to provide important information for communities and decision-makers who are facing the challenges of swift environmental change, and to offer insights for disaster mitigation planning.
Students are encouraged to email me at sankarrd@longwood.edu to discuss ongoing and potential future projects.
Amanda Starr, Ph.D.
*** Dr. Starr is not currently seeking research students. ***
Molecular Microbiology
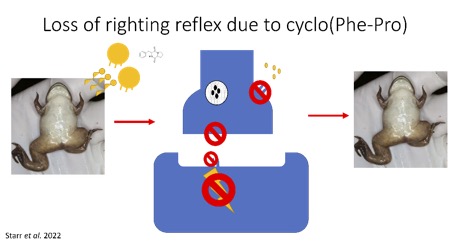
Interested in research in microbiology, genetics, chemistry/metabolomics, or ecology?? The Starr Lab explores the genetics and ecology of an invasive chytrid fungus (Batrachochytrium dendrobatidis) known to cause the decline of over 500 species of amphibians worldwide (90 species are extinct!). Current research projects are: 1) Determining minimal media conditions to culture the fungus using genome knowledge and growth curves and using that knowledge to manipulate carbon: nitrogen ratios to influence virulence and pathogenic fungal traits and 2) Studying the relationship between the amphibian bacteria-fungal relationships and their impact on fungal growth, motility, metabolite production, and germ tube development. In addition to Batrachochytrium work, the Starr Lab will be exploring antibiotic resistance in bacteria found in the air and rain. Exploration of genes of antibiotic resistance in microbes will be tested as well as understanding where these organisms came from and what this means for the current health of individuals.
Overall skills that can be learned in the Starr Lab are culturing microorganisms and learning ways to manipulate these conditions, use of basic microscopy, genetic analysis (computer and in-lab) with primer design, PCR techniques, chemical analysis, and working with DNA and RNA. There are no specific prerequisites required to be in the lab, but overall enthusiasm is encouraged. Students can add onto the above projects or new projects can be developed that focus on medical or environmental aspects of microorganisms. Get in contact with Dr. Starr (starram@longwood.edu) if you have any questions/interest.
Ben Campbell, Ph.D.
*** Dr. Campbell is not currently accepting research students.***
Science Education Research
Dr. Campbell is interested in a variety of topics in science education, ranging from instructional preparation to classroom interactions. A recent publication focused on science competitions, and another addressed opportunities for informal science education while visiting wetlands in Virginia State Parks.
